Traditional RNA detection methods, which analyze RNA levels in lysed, permeabilized, or fixed cell samples, make it challenging to understand the role of one gene in the context of a tissue, organ, or other heterogeneous cell population. Quantitative real-time polymerase chain reaction (RT-PCR), for example, reports the average quantity of RNA within a whole sample of cell. The results, while widely accepted, may not accurately reflect gene expression subpopulations within heterogeneous sample settings.
If a small subset of cells responds to a treatment or simulation by increasing transcription of a certain gene, this change could easily be masked by the rest of the population when the entire sample is lysed and its total RNA extracted. This inability to identify variations in expression levels across subpopulations can be problematic in immunology and cancer research, where efforts to understand the genetic basis of tumor progression have been greatly challenged by tumor heterogeneity and cell-extrinsic effects.
In contrast, examining RNA content in individual, live cells enables identification of smaller cell populations that have responded to a treatment differently than the rest of the cells present. Furthermore, the ability to use those same cells for downstream experiments can enable sorting and enrichment of cells based on individual RNA levels.
Our studies, performed using SmartFlare™ RNA detection probes (EMD Millipore, Boston, MA) in conjunction with flow cytometry, microscopy, and imaging cytometry, illustrate the value of live cell RNA detection at the single-cell level to identify differences in expression levels across populations of cells. In particular, we demonstrate the technique’s use in identifying cell subpopulations in live heterogeneous mixtures based on IL-6 mRNA expression, a determination that cannot be achieved through conventional RNA detection techniques.
Methods
SmartFlare technology enables specific fluorescence-based detection of RNA within live cells. The detection probes are gold nanoparticles conjugated to duplexed oligonucleotides. In the absence of target RNA, the probe is quenched by gold and does not fluoresce. In the presence of the target, the probe binds the capture strand, releasing the fluorescent reporter from the proximity of the gold and allowing fluorescence to be detected. Over time, the probes exit the cell through natural exocytosis, without adverse effects, leaving the cells intact for further analysis.
 Figure 1 ‒ Cell lysates of IL-6 negative MCF-7 and IL-6 positive MDA-MB-231 cells, along with mixtures of the two cell types in the ratios shown, were subjected to qRT-PCR analysis using an IL-6 specific primer set.
Figure 1 ‒ Cell lysates of IL-6 negative MCF-7 and IL-6 positive MDA-MB-231 cells, along with mixtures of the two cell types in the ratios shown, were subjected to qRT-PCR analysis using an IL-6 specific primer set.In this experiment, MCF-7 (Interleukin-6 [IL-6] negative) and MDA-MB-231 (IL-6 positive) human breast carcinoma cell lines were incubated with Human IL-6 Cy5 SmartFlare probe for 16 hr at 37 °C. Mixtures of the two cell lines were prepared at various MCF-7:MDA-MB-231 ratios (shown in Figure 1) and analyzed along with pure cell samples. We were aiming to simulate experimental settings where the expression of an mRNA is heterogeneous within a cell sample; MCF-7 and MDA-MB-231 breast cancer cell lines were selected because they vary significantly in their expression of IL-6 mRNA.1,2
The pure and mixed cell samples were then analyzed for IL-6 expression using three detection methods: quantitative real-time PCR (qRT-PCR), flow cytometry, and confocal microscopy. For qRT-PCR, total RNA was extracted from each cell sample, and then subjected to analysis using a TaqMan® IL-6 primer set and RNA-to-Ct™ kit (Life Technologies, Carlsbad, CA). A LightCycler® 480 real-time PCR instrument (Roche, Indianapolis, IN) was used for amplification.
For flow cytometry detection, cell samples were analyzed using a guava easyCyte™ 8HT benchtop flow cytometer (EMD Millipore). Cell populations were resolved based on IL-6 SmartFlare probe-specific Cy5 fluorescence signal. For select samples, imaging cytometry was also performed using an ImageStream®x Mark II instrument (Amnis, Seattle, WA [part of EMD Millipore]).
Finally, for confocal microscopy, cell samples were analyzed using a guava easyCyte 8HT benchtop flow cytometer scanning system with 405-nm and 640-nm argon lasers. Cells were additionally stained with 4',6-diamidino-2-phenylindole (DAPI) nuclear dye after the probe incubation.
Results
Quantitative real-time PCR
qRT-PCR is effective in determining overall levels of IL-6 mRNA in mixed cell samples. However, the method is limited in its ability to resolve subpopulations of varied expression.
In these studies, we examined lysates generated from cell samples for IL-6 expression using qRT-PCR. As expected, we observed a significant difference in expression between the pure MCF-7 cells (IL-6 negative) and MDA-MB-231 cells (IL-6 positive), with a gradient of expression levels in the mixed cell samples depending on the percentage of MDA-MB-231 cells present. However, we were not able to resolve single cells or subpopulations based on mRNA levels. Note that in Figure 1, a near 50/50 mixture of IL-6 positive and negative cell types yields a Ct value that is similar to the Ct value from a pure population of IL-6 negative cells.
To more effectively resolve single cells in the breast cancer cell mixtures based on differences in their gene expression, we analyzed the cell samples using an IL-6 Cy5 SmartFlare RNA detection probe. The technology enables relative quantification of specific RNA sequences using fluorescent detection platforms. Furthermore, treatment of cells with the probes does not compromise their integrity or viability, allowing for live cell analysis and downstream experimentation.
To assess IL-6 mRNA expression, we analyzed the probe-mediated fluorescence in the breast cancer cell samples using flow cytometry. Just as with qRT-PCR, the differences in expression levels between MCF-7 and MDA-MB-231 cell types could be clearly observed. However, while qRT-PCR only provided information about averaged IL-6 expression levels, the probe technology enabled us to study variation in expression within the mixed cell samples on a single cell level. Specifically, we observed and quantified the distribution of single cell events in the Cy5 low and Cy5 high gates in the heterogeneous cell mixtures, representative of MCF-7 and MDA-MB-231 cells, respectively, while also gaining information about overall IL-6 mRNA expression (Figure 2).
 Figure 2 ‒ Pure and mixed cell samples are subjected to flow cytometry analysis following 16-hr incubation with IL-6 Cy5 SmartFlare probes. a) (above) MCF-7 and MDA-MBA-231 cells can be clearly identified and quantified in cell samples based on RED2 channel fluorescence intensity, particularly when using a dot plot to visualize cell populations. b) (below) As the percentage of MDA-MB-231 cells increases in the mixed sample, higher overall Cy5 mean fluorescence is detected.
Figure 2 ‒ Pure and mixed cell samples are subjected to flow cytometry analysis following 16-hr incubation with IL-6 Cy5 SmartFlare probes. a) (above) MCF-7 and MDA-MBA-231 cells can be clearly identified and quantified in cell samples based on RED2 channel fluorescence intensity, particularly when using a dot plot to visualize cell populations. b) (below) As the percentage of MDA-MB-231 cells increases in the mixed sample, higher overall Cy5 mean fluorescence is detected.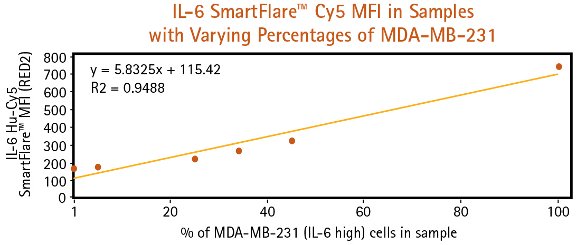
MCF-7 and MDA-MB-231 cells were subjected to confocal microscopy and imaging cytometry after incubation with IL-6 Cy5 SmartFlare probe. In the confocal micrographs, both cell types retained their normal morphologies, even in the presence of probes, and the differences in Cy5 fluorescence between the two cell types could be easily distinguished (Figure 3). In addition, imaging cytometry allowed for more in-depth correlation between qualitative and quantitative resolution of MCF-7 and MDA-MB-231 cells based on IL-6 expression.
A
 Figure 3 ‒ a) MCF-7 and MDA-MB-231 cells were incubated with IL-6 Cy5 SmartFlare probes for 16 hr, stained with Hoechst 33342 DNA dye, and subjected to confocal microscopy. The two cell types can be distinguished both by morphological differences as well as distinctive Cy5 fluorescence relative to their IL-6 mRNA expression. b) When analyzing a mixed MCF-7/MDA-MB-231 cell sample using an ImageStreamx Mark II imaging cytometer, one can correlate subpopulations of high and low Cy5 fluorescence with images of individual cells contained within these populations.
Figure 3 ‒ a) MCF-7 and MDA-MB-231 cells were incubated with IL-6 Cy5 SmartFlare probes for 16 hr, stained with Hoechst 33342 DNA dye, and subjected to confocal microscopy. The two cell types can be distinguished both by morphological differences as well as distinctive Cy5 fluorescence relative to their IL-6 mRNA expression. b) When analyzing a mixed MCF-7/MDA-MB-231 cell sample using an ImageStreamx Mark II imaging cytometer, one can correlate subpopulations of high and low Cy5 fluorescence with images of individual cells contained within these populations.Summary
In this study, we have demonstrated the use of fluorescent detection technology to identify cell subpopulations in live heterogeneous mixtures based on IL-6 mRNA expression. qRT-PCR was able to detect IL-6 mRNA in breast cancer cell lines, but could not distinguish between high- and low-expressing subpopulations in heterogeneous cell samples. In contrast, the SmartFlare probes could detect mRNA while also identifying and resolving subpopulations based on gene expression. We also showed the RNA detection technology could be used to study gene expression without compromising the viability of individual cells. This capability enabled downstream experimentation and correlation of gene expression to cell behavior.
Many focus areas of cell biology, such as immunology and cancer research, require the resolution of cell subpopulations within heterogeneous samples based on gene expression levels. By allowing researchers to specifically detect RNA in single, live cells—and therefore identify variations in expression levels—the new RNA probes help generate more biologically significant gene expression data.
References
- Lee, S.T.; Li, Z et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 2011, 43, 798‒810.
- Sasser, A.K.; Sullivan, M.J. et al. Interleukin-6 is a potent growth factor for ER-α-positive human breast cancer. FASEB J. Res. Comm. 2007, 21, 3763‒70.
Victor Koong is Product Manager; Alex Ko, Yuko Williams, and Laura White are Research Scientists; and Don Weldon is R&D Manager, EMD Millipore, 17 Cherry Hill Dr., Boston, MA 01923, U.S.A.; tel.: 978-762-5324; e-mail: [email protected] ; www.emdmillipore.com